Time Lapse Registration
Time lapse registration is necessary because the imaged specimen may, and in most cases will, drift over a long acquisition period. In OpenSPIM this situation is typically exaggerated because:
a) we set-up the imaging angles quickly and approximately, relying on later re-adjustment based on imaging result (thus the first time-points are usually quite off with respect to the rest of the time lapse - see sample raw data),
b) the motors precision is limited.
The pre-requisite for time-series registration is to perform all individual time-point registrations, particularly the segmentation of the beads.
Note that it is possible to do time-series registration also directly, i.e. segment the beads and do time-series registration in one step, however this is not a typical workflow.
As before we walk through the set-up of plugin input, annotate the output and look into the new registration files created.
Input
|
|
We start by going back to registration plugin, Plugins->SPIM registration->Bead-based registration, and keep the same parameters as before in the first dialog (Single channel and Difference-of-Mean). |
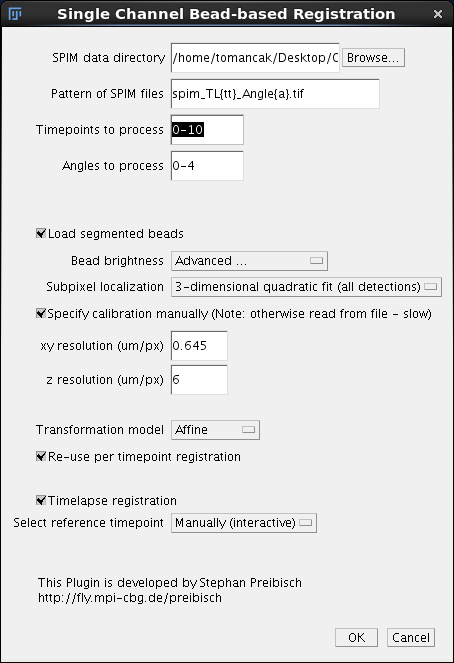
The main dialog is the same as the one we used in individual time-point registration. The top set of parameters specifying file location and format should be pre-filled. We need to only change the Timepoints to process to 0-10. Next we click the checkbox Load segmented beads which signifies that the plugin should simply look into the previously established .beads.txt files in the /registration subfolder and load them. This will happen very fast compared to re-segmenting the beads. All parameters for bead segmentation are ignored. We need to specify the calibration as explained before. We will use the affine transformation model. The option Re-use per time point registration should be clicked. I am actually not sure what it does, but I assume that it will load the transformation models determined in the per time-point registration and use them as a starting point. Finally, we must decide how will the reference time-point be determined. Reference time-point is the time-point to which all others will be registered, see here for detailed explanation. We have three option for how to choose it:
We choose Manually (interactive) and continue by pressing OK. |
|
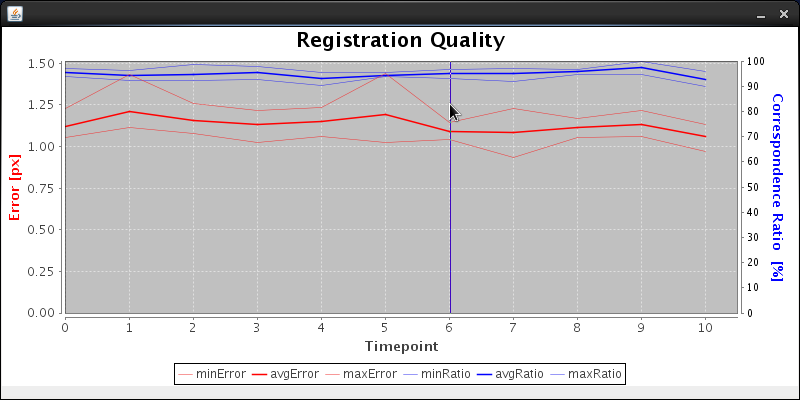 |
There is some output in the Log window which we will discuss momentarily. Then a new window pops up summarizing the registration errors of the individual time-point registrations. Click with the mouse on the time-point which you want to choose as reference. Good options are :
Note that for long time-series where the specimen changes size, the registration of time-points far away from the reference may be suboptimal. In that case we recommend splitting the time-series into several independent segments and register separately to the middle time-point of each segment. The entire time series can then be stablized using Fiji's Descriptor based registration. The description of this complex situation goes beyond the scope of this tutorial. We click on time-point number 6 and the time-series registration commences. |
Run
The output of time lapse registration rolls by quickly through the Log window. All we are doing is read text files (bead positions) and running the registration, which as we saw before, is quick. The entire output can be downloaded here.
Version 0.55
(Mon Jun 17 15:31:22 CEST 2013): Starting Bead Extraction
Read 1275 beads for spim_TL00_Angle0.tif (id = 0)
Read 1364 beads for spim_TL00_Angle1.tif (id = 1)
Read 1026 beads for spim_TL00_Angle2.tif (id = 2)
Read 1110 beads for spim_TL00_Angle3.tif (id = 3)
Read 1201 beads for spim_TL00_Angle4.tif (id = 4)
(Mon Jun 17 15:31:22 CEST 2013): Finished Bead Extraction
(Mon Jun 17 15:31:22 CEST 2013): Starting Registration
(Mon Jun 17 15:31:22 CEST 2013): Finished Registration
Finished processing.
(Mon Jun 17 15:31:22 CEST 2013): Starting Bead Extraction
Read 1243 beads for spim_TL01_Angle0.tif (id = 0)
Read 1467 beads for spim_TL01_Angle1.tif (id = 1)
Read 970 beads for spim_TL01_Angle2.tif (id = 2)
Read 1125 beads for spim_TL01_Angle3.tif (id = 3)
Read 1172 beads for spim_TL01_Angle4.tif (id = 4)
(Mon Jun 17 15:31:22 CEST 2013): Finished Bead Extraction
(Mon Jun 17 15:31:22 CEST 2013): Starting Registration
(Mon Jun 17 15:31:23 CEST 2013): Finished Registration
Finished processing.
.
.
First we load the previously segmented beads and execute the per time-point registration (this is done solely for the purpose of getting the data for the graph of registration errors). After that the graphical overview of registration errors pops up as described above. Clicking time-point 6 triggers the next barrage of output:
(Mon Jun 17 15:37:43 CEST 2013): Loading reference timepoint information Reference ViewStructure Timepoint 6
Read 1012 beads for spim_TL06_Angle0.tif (id = 0)
Read 1169 beads for spim_TL06_Angle1.tif (id = 1)
Read 950 beads for spim_TL06_Angle2.tif (id = 2)
Read 1008 beads for spim_TL06_Angle3.tif (id = 3)
Read 1153 beads for spim_TL06_Angle4.tif (id = 4)
(Mon Jun 17 15:37:43 CEST 2013): Loading timepoint Template ViewStructure Timepoint 0 information
Read 1275 beads for spim_TL00_Angle0.tif (id = 0)
Read 1364 beads for spim_TL00_Angle1.tif (id = 1)
Read 1026 beads for spim_TL00_Angle2.tif (id = 2)
Read 1110 beads for spim_TL00_Angle3.tif (id = 3)
Read 1201 beads for spim_TL00_Angle4.tif (id = 4)
Once again the beads are loaded from files in /registration folder, this time for the time-point at hand and the reference time-point.
spim_TL00_Angle2.tif<->spim_TL00_Angle3.tif: Remaining inliers after RANSAC: 27 of 27 (100%) with average error 0.6691474128100607
spim_TL00_Angle2.tif<->spim_TL00_Angle4.tif: Remaining inliers after RANSAC: 37 of 38 (97%) with average error 0.44379338863733653
spim_TL06_Angle0.tif<->spim_TL00_Angle2.tif: Remaining inliers after RANSAC: 14 of 15 (93%) with average error 0.7140956042068345
spim_TL00_Angle1.tif<->spim_TL00_Angle3.tif: Remaining inliers after RANSAC: 52 of 55 (95%) with average error 0.6072635080378788
spim_TL06_Angle1.tif<->spim_TL00_Angle2.tif: Remaining inliers after RANSAC: 14 of 14 (100%) with average error 0.6171177732092994
spim_TL00_Angle1.tif<->spim_TL00_Angle4.tif: Remaining inliers after RANSAC: 32 of 35 (91%) with average error 0.4623754050116986
spim_TL00_Angle0.tif<->spim_TL00_Angle2.tif: Remaining inliers after RANSAC: 35 of 38 (92%) with average error 1.016280529328755
spim_TL06_Angle0.tif<->spim_TL00_Angle1.tif: Not enough correspondences found 4, should be at least 12
spim_TL06_Angle2.tif<->spim_TL00_Angle0.tif: Remaining inliers after RANSAC: 17 of 18 (94%) with average error 0.6060983157333206
spim_TL06_Angle0.tif<->spim_TL00_Angle3.tif: Remaining inliers after RANSAC: 25 of 27 (93%) with average error 0.850523499250412
spim_TL06_Angle1.tif<->spim_TL00_Angle4.tif: Remaining inliers after RANSAC: 18 of 18 (100%) with average error 0.6293857701950603
spim_TL06_Angle1.tif<->spim_TL00_Angle3.tif: Remaining inliers after RANSAC: 34 of 34 (100%) with average error 0.6525622424395645
spim_TL00_Angle3.tif<->spim_TL00_Angle4.tif: Remaining inliers after RANSAC: 42 of 43 (98%) with average error 0.8881665209219568
spim_TL06_Angle2.tif<->spim_TL00_Angle1.tif: Remaining inliers after RANSAC: 13 of 15 (87%) with average error 0.3753714802173468
spim_TL06_Angle1.tif<->spim_TL00_Angle0.tif: Remaining inliers after RANSAC: 16 of 17 (94%) with average error 1.0114890160039067
ViewErrorStatistics.setViewSpecificError(): Warning! spim_TL00_Angle1.tif (id = 6) is not part of the structure of my view spim_TL06_Angle0.tif (id = 0)
spim_TL00_Angle0.tif<->spim_TL00_Angle3.tif: Remaining inliers after RANSAC: 51 of 55 (93%) with average error 0.8029027958126628
spim_TL00_Angle1.tif<->spim_TL00_Angle2.tif: Remaining inliers after RANSAC: 37 of 37 (100%) with average error 0.6356905201399647
spim_TL00_Angle0.tif<->spim_TL00_Angle4.tif: Remaining inliers after RANSAC: 42 of 44 (95%) with average error 0.8402975451733384
spim_TL06_Angle0.tif<->spim_TL00_Angle4.tif: Remaining inliers after RANSAC: 24 of 24 (100%) with average error 0.4517987013484041
spim_TL00_Angle0.tif<->spim_TL00_Angle1.tif: Remaining inliers after RANSAC: 40 of 42 (95%) with average error 1.0302402991801498
spim_TL06_Angle2.tif<->spim_TL00_Angle2.tif: Remaining inliers after RANSAC: 100 of 102 (98%) with average error 0.21032628208398818
spim_TL06_Angle1.tif<->spim_TL00_Angle1.tif: Remaining inliers after RANSAC: 80 of 83 (96%) with average error 0.21069390852935613
spim_TL06_Angle2.tif<->spim_TL00_Angle3.tif: Remaining inliers after RANSAC: 18 of 22 (82%) with average error 0.45325933520992595
spim_TL06_Angle0.tif<->spim_TL00_Angle0.tif: Remaining inliers after RANSAC: 81 of 82 (99%) with average error 0.21133165388011638
spim_TL06_Angle4.tif<->spim_TL00_Angle1.tif: Remaining inliers after RANSAC: 15 of 16 (94%) with average error 0.6895617981751759
spim_TL06_Angle4.tif<->spim_TL00_Angle2.tif: Remaining inliers after RANSAC: 39 of 41 (95%) with average error 0.6350838870574267
spim_TL06_Angle4.tif<->spim_TL00_Angle0.tif: Remaining inliers after RANSAC: 13 of 14 (93%) with average error 0.6161291100657903
spim_TL06_Angle3.tif<->spim_TL00_Angle0.tif: Remaining inliers after RANSAC: 16 of 18 (89%) with average error 0.7691099494695663
spim_TL06_Angle4.tif<->spim_TL00_Angle3.tif: Remaining inliers after RANSAC: 17 of 17 (100%) with average error 0.5228452130275614
spim_TL06_Angle2.tif<->spim_TL00_Angle4.tif: Remaining inliers after RANSAC: 36 of 37 (97%) with average error 0.41419285929037464
spim_TL06_Angle3.tif<->spim_TL00_Angle4.tif: Remaining inliers after RANSAC: 17 of 19 (89%) with average error 0.47992865653599
spim_TL06_Angle3.tif<->spim_TL00_Angle2.tif: Remaining inliers after RANSAC: 24 of 26 (92%) with average error 0.552938544501861
spim_TL06_Angle3.tif<->spim_TL00_Angle1.tif: Remaining inliers after RANSAC: 46 of 47 (98%) with average error 0.44524666051501816
spim_TL06_Angle3.tif<->spim_TL00_Angle3.tif: Remaining inliers after RANSAC: 116 of 120 (97%) with average error 0.3821218167913371
spim_TL06_Angle4.tif<->spim_TL00_Angle4.tif: Remaining inliers after RANSAC: 144 of 144 (100%) with average error 0.21463561007597798
spim_TL06_Angle0.tif (id = 0) has 144 correspondences in 4 other views.
spim_TL06_Angle1.tif (id = 1) has 162 correspondences in 4 other views.
spim_TL06_Angle2.tif (id = 2) has 184 correspondences in 4 other views.
spim_TL06_Angle3.tif (id = 3) has 219 correspondences in 4 other views.
spim_TL06_Angle4.tif (id = 4) has 228 correspondences in 4 other views.
spim_TL00_Angle0.tif (id = 5) has 311 correspondences in 4 other views.
spim_TL00_Angle1.tif (id = 6) has 315 correspondences in 4 other views.
spim_TL00_Angle2.tif (id = 7) has 327 correspondences in 4 other views.
spim_TL00_Angle3.tif (id = 8) has 382 correspondences in 4 other views.
spim_TL00_Angle4.tif (id = 9) has 392 correspondences in 4 other views.
The total number of detections was: 0
The total number of true correspondences is: 1332
The total number of correspondence candidates was: 1388
The ratio is: 96%
The 10 views are thrown together in one pile and correspondences for all pair-wise combinations are established. See here for more detailed discussion. Most view pairs have correspondences apart from TL00_Angle1 and TL06_Angle0. Altogether the two sets of angles across two timepoints are tied together by 1332 correspondences and the RANSAC inlier ratio is excellent - 96%. Note that this is particularly possible for Drosophila which does not grow across time displacing the beads.
Fixing tile spim_TL06_Angle0.tif (id = 0)
Fixing tile spim_TL06_Angle1.tif (id = 1)
Fixing tile spim_TL06_Angle2.tif (id = 2)
Fixing tile spim_TL06_Angle3.tif (id = 3)
Fixing tile spim_TL06_Angle4.tif (id = 4)
Successfully optimized configuration of 10 tiles after 210 iterations:
average displacement: 1.025px
minimal displacement: 0.861px
maximal displacement: 1.243px
Optimizer Matrices
spim_TL06_Angle0.tif (id = 0):
Transformation:
3d-affine: (1.0, 0.0, 0.0, 0.0, 0.0, 1.0, 0.0, 0.0, 0.0, 0.0, 1.0, 0.0)
Scaling: (1.0, 1.0, 1.0)
spim_TL06_Angle1.tif (id = 1):
Transformation:
3d-affine: (0.4019471, -0.018123448, -0.94941515, 643.9887, -0.026565894, 0.99999356, -0.031975985, 86.95143, 0.90984946, 0.015314827, 0.32108918, -478.69415)
Scaling: (1.0024096594589158, 0.9508485621397472, 1.0426960902620872)
spim_TL06_Angle2.tif (id = 2):
Transformation:
3d-affine: (-0.74307096, -0.04229408, -0.6503768, 1350.867, -0.045259617, 0.99987435, -0.00910145, 107.2758, 0.62513214, 0.002267208, -0.805896, -41.837677)
Scaling: (0.969682848632148, 1.0391979831248561, 0.9994296423258292)
spim_TL06_Angle3.tif (id = 3):
Transformation:
3d-affine: (-0.8651256, -0.037340384, 0.5560009, 1292.4794, -0.032303065, 0.99945796, 0.004552126, 81.7959, -0.5200823, -0.025575923, -0.82603866, 843.71893)
Scaling: (1.0296682501585668, 0.9743926675610988, 1.0013869457729445)
spim_TL06_Angle4.tif (id = 4):
Transformation:
3d-affine: (0.50565606, -0.0051612062, 0.8625701, 122.84003, -8.2300743E-4, 1.0010158, -0.0058299266, 79.84306, -0.8234671, -0.023073934, 0.56407094, 650.40784)
Scaling: (0.9624436529435839, 1.0379529258315343, 0.9974826010869479)
spim_TL00_Angle0.tif (id = 5):
Transformation:
3d-affine: (0.99880403, -2.428703E-4, -0.0041077733, 61.020546, -1.813916E-4, 0.999382, 0.0040932633, 358.4858, 0.0036993474, 8.380413E-5, 0.9987223, 21.614502)
Scaling: (0.9987749991624741, 1.001208654485056, 0.9969439914051773)
spim_TL00_Angle1.tif (id = 6):
Transformation:
3d-affine: (0.3984493, -0.017852351, -0.9511509, 614.63696, -0.025950752, 0.9992613, -0.030624092, 271.6453, 0.9109962, 0.014979344, 0.32206786, -418.2312)
Scaling: (0.9535989906105291, 1.0016059697570685, 1.0417310799511799)
spim_TL00_Angle2.tif (id = 7):
Transformation:
3d-affine: (-0.7449159, -0.042260386, -0.64679694, 1300.0942, -0.04498928, 0.9988398, -0.009780109, 240.2511, 0.6214917, 0.0016765222, -0.80720603, -89.87463)
Scaling: (0.9688138080167756, 1.0378264989295503, 0.9984912630908743)
spim_TL00_Angle3.tif (id = 8):
Transformation:
3d-affine: (-0.8629845, -0.03721965, 0.5713175, 1311.1292, -0.032652352, 0.99901617, 0.012039125, 219.21135, -0.52140266, -0.025454005, -0.8308716, 768.1265)
Scaling: (1.0380281150297346, 1.0031215112339598, 0.9751290665644732)
spim_TL00_Angle4.tif (id = 9):
Transformation:
3d-affine: (0.5076602, -0.0047055706, 0.86028206, 70.25545, -0.0016549313, 0.9997853, 0.003312394, 149.69363, -0.82133454, -0.023289602, 0.56215394, 807.2336)
Scaling: (0.961987733549366, 1.0320840492530776, 0.9989707811248745)
Finally the global optimization is run and we get the transformation matrices for all 10 views involved. The Angle0 of TL06 serves as a reference frame to which all other time-points are registered. Note that all angles get new transformation matrices since they are now registered in the context of a global optimization run on 10 angles (5 reference and 5 time-point specific).
This process is repeated for every time-point in the time series, i.e. 11 times for the sample data.
Output
The output is, similarly to individual time-point registration, a series of text files in the /registration directory.
cd registration/
ls
spim_TL00_Angle0.tif.beads.txt
spim_TL00_Angle0.tif.dim
spim_TL00_Angle0.tif.registration
spim_TL00_Angle0.tif.registration.to_6
spim_TL00_Angle1.tif.beads.txt
spim_TL00_Angle1.tif.dim
spim_TL00_Angle1.tif.registration
spim_TL00_Angle1.tif.registration.to_6
spim_TL00_Angle2.tif.beads.txt
spim_TL00_Angle2.tif.dim
spim_TL00_Angle2.tif.registration
spim_TL00_Angle2.tif.registration.to_6
.
.
.
The new files end with the suffix .to_6 indicating that these are time-lapse registration files to the reference time-point number 6. The content of the files is equivalent to the .registration file discussed in the registration section, however the matrices are different.
We will now proceed to fuse the data using this new time lapse registration and compare the output to the individual per time-point registrations.
Fusion of time lapse data
We have seen in the fusion part of the tutorial how it is necessary to first run fusion on downsampled data, define the cropping area and then re-run the pipeline with full resolution data. To simplify, we will skip here the steps required for defining the cropping area. It is important to note that this has to be done again as we need to define the crop area relative to the new time lapse registration. Below we show how to run the fusion on the time lapse registered data at full resolution across all time-points.
|
|
Back to Plugins->SPIM registration->Multi-view fusion. First dialog remains the same (pre-loaded). |
In the next dialog we will modify the Time point to process to 0-10. |
|
In the main dialog the important thing to remember is to choose the correct registration file. In the first pull down menu Registration for channel 0 select Time-point registration (reference=6) of channel 0. The remaining parameters remain the same (for details see fusion). Note that the cropping parameters were determined by running the fusion with the new to time-point 6 reference registration and recording the crop area coordinates as described before. |
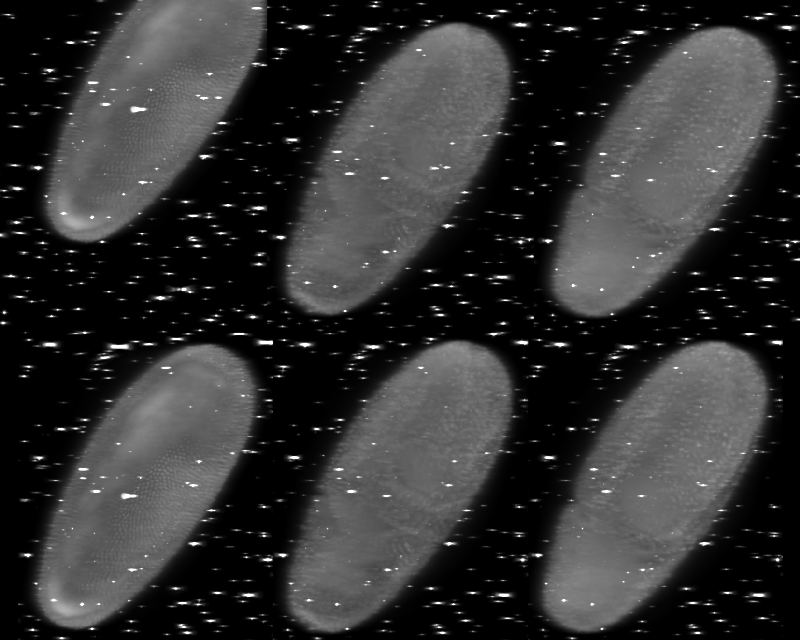
Let's now examine how the time-lapse registration changed the output. As we saw in the raw data the first angle of the first time point was way off the field of view. If we do not apply the time lapse registration, the first time-point will be registered but shifted with respect to the rest of the time-lapse (see picture - top row). After the time lapse registration the embryos are well aligned across time-points (bottom row).
We have now FINISHED the SPIMage processing pipeline. We registered all time points across space and time and we fused them all at full resolution. The results are in /registration and /output directories. On this page we will learn how to browse the raw, registered and fused data in various ways.