Bryozoan and Brachiopod development
Imaging live bryozoan embryos
Shipping live colonies
Colonies of Membranipora membranacea needed to be shipped from Bergen, Norway to Dresden, Germany. The first package contained around 10 pieces of kelp blades with colonies inside single transparent plastic bag filled with sea water (bubbled overnight) and sealed with tape. Bag was surrounded by icepacks to keep temperatures low. Upon arrival we realized that the plastic bag had ruptured during the trip and the colonies were out of the water for an unknown period of time. They were placed in a 50 L plastic tank in the cold room (4 °C) immediately. After 1h I checked under the scope and all the colonies were dead.
Our colleagues in Bergen prepared an emergency second shipment, this time with colonies placed inside three plastic bags with sea water (not bubbled) and less air to avoid water movement that could rip the plastic. Upon arrival two of the inner plastic bags were broken, but colonies were still underwater. They were split in two groups to be kept at 4 °C and 12 °C and were acclimatized in 1:1 natural:artificial sea water for 1h. Then they were transferred to artificial sea water. Under the scope zooids were still moving. On the following morning, however, most of the colonies were dead or just-dead, but still with some normal embryos which I extracted as described below.
Acknowledgements: Annie Boddington and Aina Børve for collection and shipping from Bergen. Yu-Wen Hsieh and Daniele Soroldoni for preparing cultures and receiving the bryozoans in Dresden.
Sample preparation
I collected fertilized eggs by scrapping the surface of ripe zooids. Eggs were activated with 0.1 mM EDTA at 12 °C for 2 hours. The few embryos that started cleaving were transferred to a 4-well plate with 5µg/mL FM 4-64 solution in sea water. After 15 min I put the embryos in a round coverslip coated with poly-l-lysine and mounted on the sample holder of the Multi-modal scanned lightsheet SPIM (Meyers SPIM). I chose this scope because it has temperature control in the chamber (15 °C) and is adapted to small samples.
Imaging
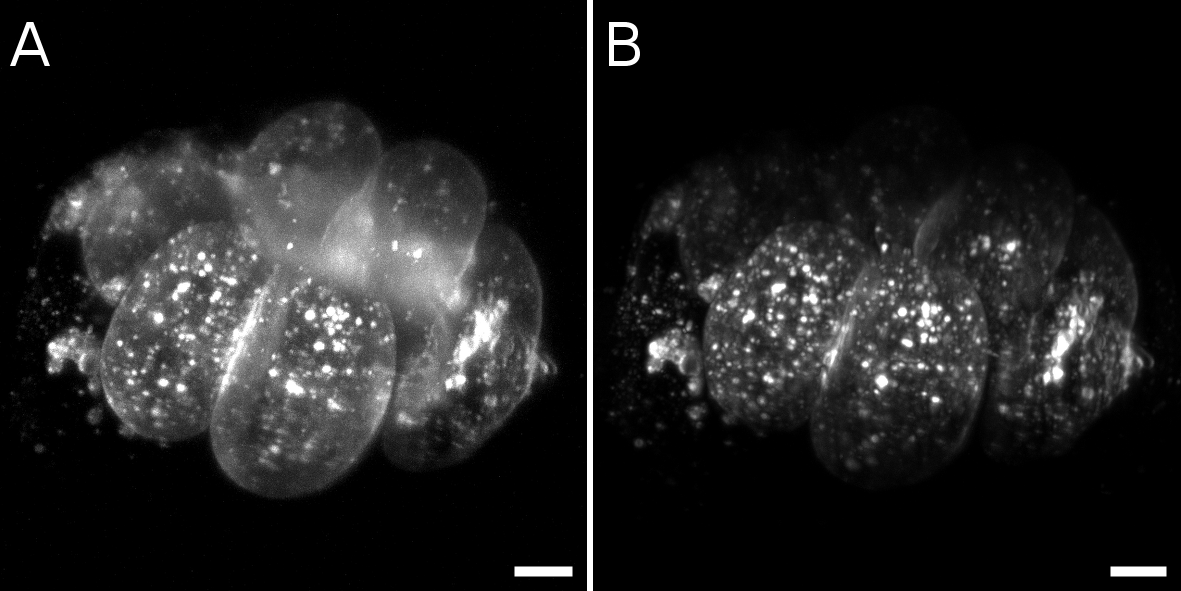
Slice-by-slice comparison between gaussian beam without (A) and with (B) structured illumination in a live bryozoan embryo. Scale bars = 10 µm
Live bryozoan embryo under Scanned Lightsheet SPIM. 201 planes were acquired every 10s for 48 timepoints and excited with 515 nm laser. Z-stacks were focused with Fiji>Time Lapse> Gaussian-based stack focuser.
Unfortunately, we only managed to acquire a small time-lapse from an embryo that was not healthy.
Acknowledgements: Loïc Royer and Nicola Maghelli for helping with the mounting and driving the scope.
Fixed bryozoan embryos and larvae
Multi-modal scanned lightsheet SPIM (Myers SPIM)
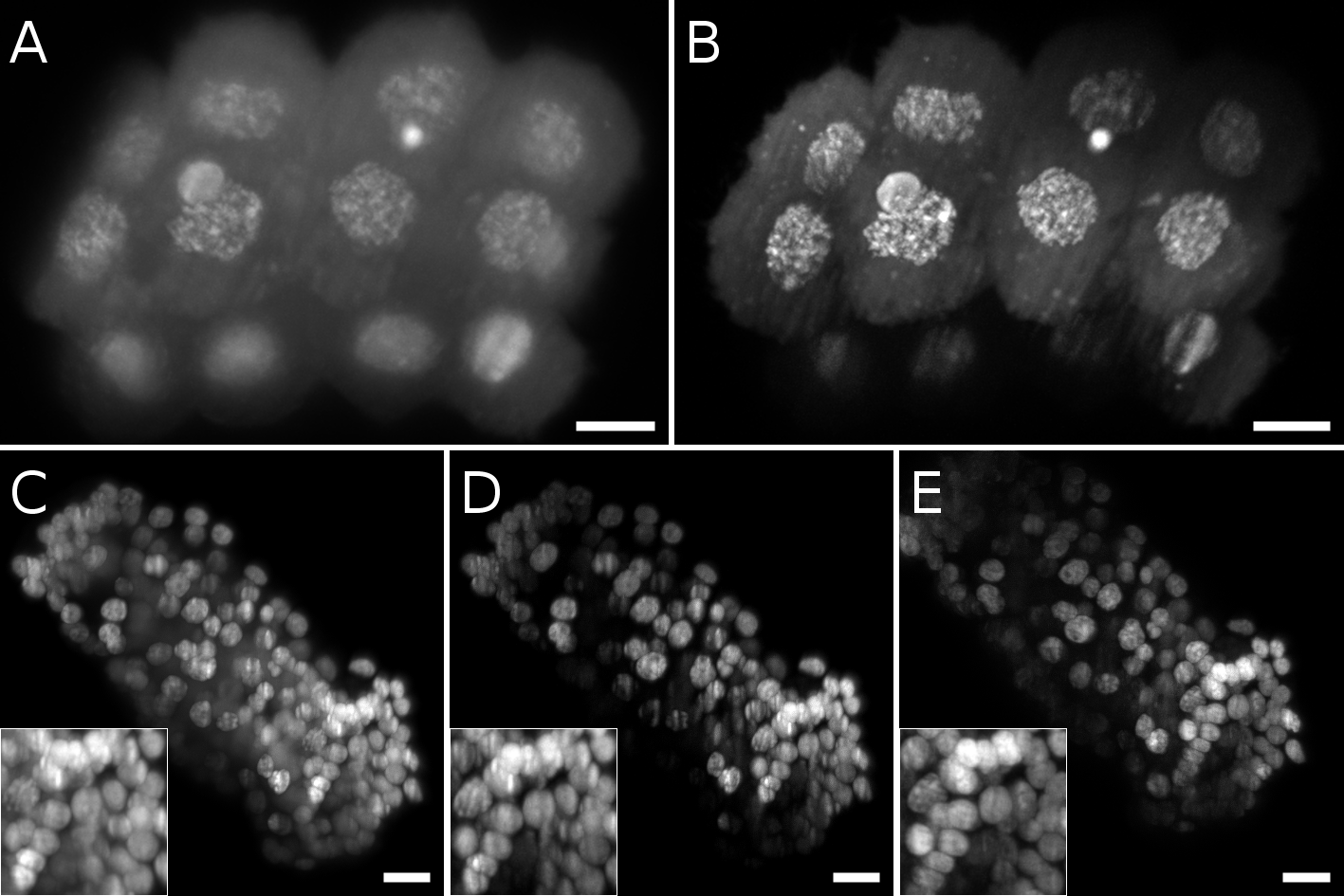
Comparison of different laser patterns in fixed bryozoan embryos. A, B Maximum intensity projection of a 16 cell stage embryo imaged with gaussian beam without (A) and with (B) structured illumination. C, D, E Maximum intensity projection of a late gastrula stage. Insets show a detail of the nuclei. C Gaussian beam. D Gaussian beam and structured illumination. E Bessel beam and structured illumination. Scale bars = 10 µm
Maximum intensity projections of two cyphonautes larvae of a bryozoan under the Multi-modal scanned lightsheet SPIM. A, D Nuclei stained with Sytox Green. B, E Actin stained with Phalloidin Alexa647. C, F Merge of the correspondent images to the left showing the nuclei (magenta) and actin (green). Scale bars = 10 µm"
Acknowledgements: Loïc Royer and Nicola Maghelli for helping with the mounting and driving the scope.
Zeiss Lightsheet Z.1
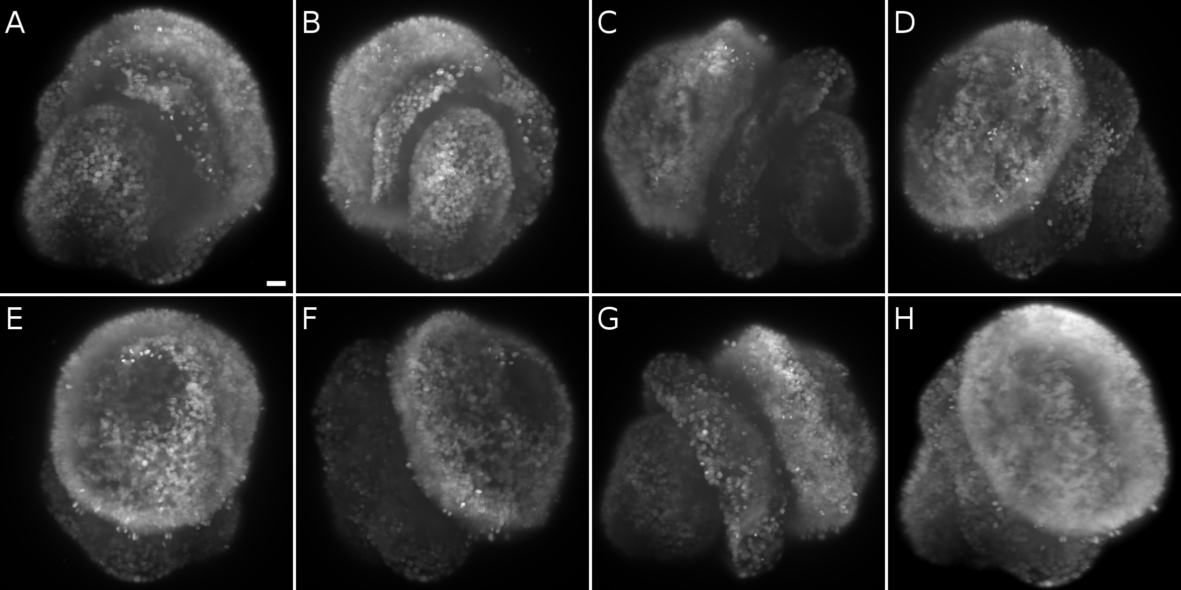
Multiview acquisition of a bryozoan larva (cyphonautes) with a nuclear (sytox green in magenta) and actin (phalloidin in green) staining. A Angle 0°. B Angle 51°. C Angle 103°. D Angle 154°. E Angle 206°. F Angle 257°. G Angle 309°. H Seven views fused with a content-based registration of both channels. Scale bar = 10 µm.
Visualization of multiple views of a bryozoan larva during registration steps. Each angle has a different color viewed on the BigDataViewer. A Original orientation of the multiple view stacks. In order to facilitate the content-based registration (without beads) the views need to be at the same orientation. A transformation was applied to the Y axis to place each view at the same orientation. B Multiple views after transformation. All stacks are oriented in the same manner. C Result of the registration step with the different views combined into a single fused stack.
Acknowledgements: Stephan Preibisch for the help with the Multiview Registration and Tobias Pietzsch for the help with the BigDataViewer.
Fixed brachiopod larvae
Zeiss Lightsheet Z.1
Multiview acquisition of a brachiopod larva with a nuclear staining (sytox green). A Angle 0°. B Angle 51°. C Angle 103°. D Angle 154°. E Angle 206°. F Angle 257°. G Angle 309°. H Seven views fused with a content-based registration of both channels. Scale bar = 10 µm.
Visualization of multiple views of a brachiopod larva during registration steps. Each angle has a different color viewed on the BigDataViewer. A Original orientation of the multiple view stacks. In order to facilitate the content-based registration (without beads) the views need to be at the same orientation. A transformation was applied to the Y axis to place each view at the same orientation. B Multiple views after transformation (left). All stacks are oriented in the same manner and have been colored individually (right). C Result of the registration step with the different views combined into a single fused stack.
Acknowledgements: Stephan Preibisch for the help with the Multiview Registration and Tobias Pietzsch for the help with the BigDataViewer. Jaroslav Icha for the help in the image acquisition and processing with the Z.1.